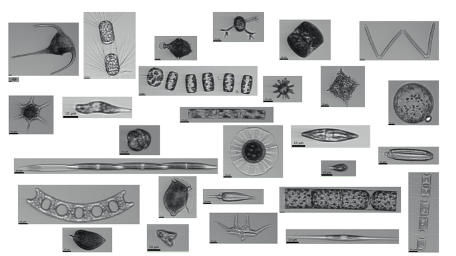
Thanks to recent advances in instrumentation, we can now observe phytoplankton – the single-celled autotrophs that form the base of the marine food web – using automated, high-throughput microscopy. Millions of images of microscopic phytoplankton cells have been collected from oceans and seas across the globe, using an instrument called the Imaging FlowCytobot (IFCB). The IFCB is deployed onboard oceanographic research vessels and captures thousands of individual particle images every hour.
Use of novel plankton imagery data to address a wide range of oceanographic and marine ecosystem questions is currently limited by the time required to analyze and categorize images. Processing these images for use in oceanographic research is time consuming, and the quantity of data necessitates the use of automated processes to classify images. Thus, the need for open-source, efficient, and effective classification tools is high. The primary objective of our project, titled User-Friendly Tools for Oceanic Plankton Image Analysis (UTOPIA) is to develop machine learning methods and code for IFCB image data classification. We aim to produce open-source, user-friendly tools - namely code to train deep learning CNNs, as well as trained networks - that allow for broad application within the oceanographic research community.
What is the role of computation in addressing the problem, and what is the nature of the computational approach?
The datasets of plankton cell imagery consist of millions of individual images (files). We use deep learning to train convolutional neural networks to categorize images based on their taxonomic classification (think machine learning image recognition but for phytoplankton cells in the ocean). Small, “toy” datasets are able to be trained on local computing resources. However, to improve the accuracy while including a sufficient number of categories to make the networks applicable to realistic, open-ocean datasets, large datasets must be used during network training. These datasets of millions of images require computing resources beyond what is do-able on local machines. Therefore, our approach was to use a Virtual Machine set up on Microsoft Azure to run python code via a JupyterLab to train our convolutional neural network. We tested pointing to Microsoft Azure blob storage data, as well as uploading the data directly to the Virtual Machine disk.
How did using cloud services advance your research?
Azure Cloud access advanced our research as the computational power we required was otherwise not available. We were able to train a dataset using our full plankton image dataset, which was an invaluable step forward in the UTOPIA project that is an ongoing effort. The collaboration value of the UTOPIA project is significant. Thus far through informal conversations with researchers who collect plankton imagery data from several different universities, there is a strong interest in these types of open-source code and tools being available. Cloud computing resources are necessary to train networks iteratively, and especially following the addition of new data.
What was your experience with the technical side of working in the cloud?
We set up our own GPU instance using the Data Science Ubuntu image, and accessed it through the built-in JupyterHub. This process was rather smooth. Reading data directly from cloud storage during deep learning training was slow, so we moved the data to the disk of the instance and thus achieved reasonable speed up in the performance. In general, we found that while the Azure website was not always completely intuitive or clear to navigate, a search for what we were trying to do nearly always resulted in a clear answer provided in the Microsoft documentation websites.
Related references:
- CHASING OPEN-OCEAN EDDIES AND FRONTS USING IN-LINE OPTICAL MEASUREMENTS (includes a section of the poster about the UTOPIA project), Ocean Optics XXV Conference, October 2-7, 2022, https://oceanopticsconference.org/poster-chase/
- Ongoing development of the UTOPIA GitHub organization: https://github.com/ifcb-utopia
Authors:
Ali Chase, Postdoctoral scholar, Applied Physics Laboratory at the University of Washington
Valentina Staneva, Senior Data Scientist, eScience Institute at the University of Washington

